At the Peixoto Lab, which I joined in June 2021, I have conducted research focused on how sleep deprivation and subsequent recovery sleep affect the transcriptome of the mouse cortex. This work aimed to deepen our understanding of the relationship between sleep disturbances and the severity of neurodevelopmental disorders, such as autism.
In this endeavor, I have employed bioinformatics tools like Salmon, various R/Bioconductor project packages, and the DAVID bioinformatics resource for RNA-seq data processing, integration, gene expression analysis, and functional enrichment analysis. I have gained experience with reproducible genomics pipelines and executing tasks on a high-performance computing cluster in a UNIX-like environment.
I completed the data analysis phase of this project, submitted samples to GEO, and am currently working to finalize figures and a manuscript for publication.
I showcased a part of my research by presenting a poster at the 2022 ISMB Conference organized by the International Society of Computational Biology.
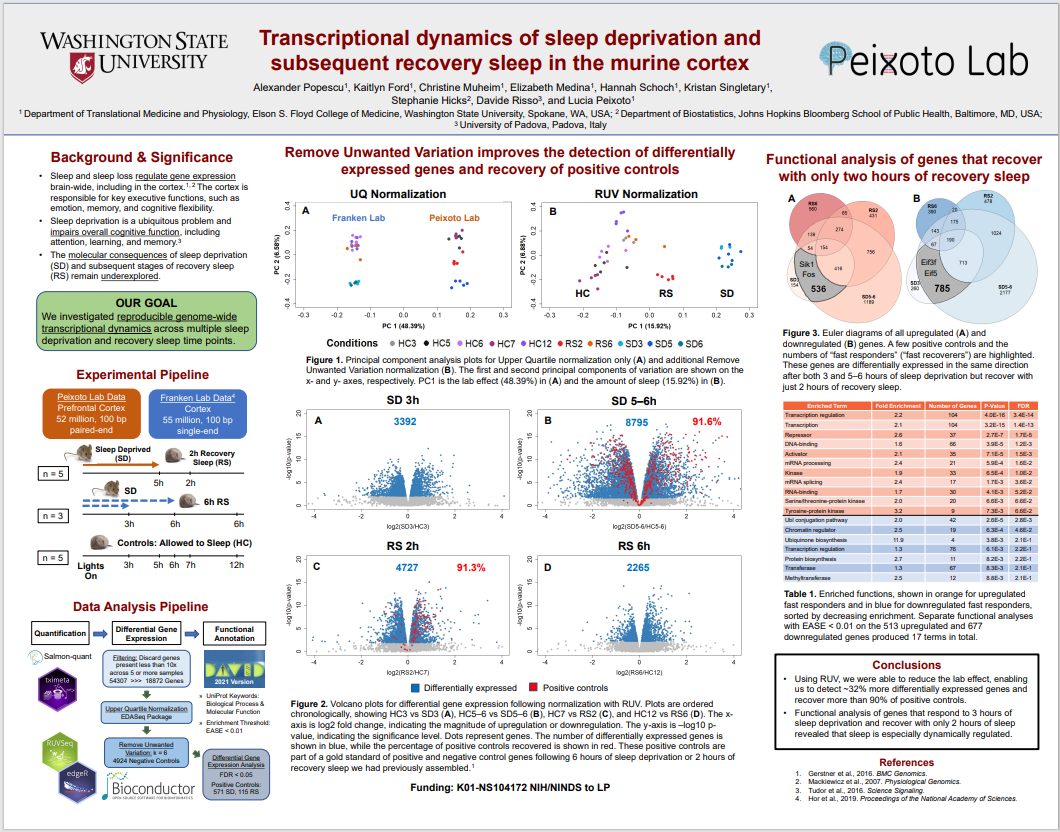
Alex’s poster from ISMB 2022

Alex with fellow members of the Peixoto Lab at ISMB 2022
Through my work with miRcore, I have gained proficiency in generating, handling, and analyzing genomics data using tools like fastq-dump, cutadapt, and bowtie2 in a UNIX command-line interface and using R. I’ve also become comfortable with online websites such as NCBI, GEO2R, STRING, KEGG, and the UCSC Genome Browser.
Through miRcore’s volunteer program, I accumulated over 100 volunteer hours and contributed towards miRcore’s aim of democratizing medical research by raising money for microgrants for early career scientists. As part of this effort, I founded a Genes in Symptoms and Diseases (GIDAS) Club at Stanford Online High School.
I collaborated with other high school students on computational biology research for opioid addiction and COVID-19, and our work Unique tRNA Fragment Upregulation with SARS-CoV-2 but Not with SARS-CoV Infection was recently published in the International Journal of Molecular Sciences.

After reviewing basic science research connecting Vitamin K2 (VK2) to factors implicated in Alzheimer’s pathogenesis as part of my “Writing in the Sciences” course at Harvard Extension School, I coauthored the first-ever literature review on VK2’s potential role in Alzheimer’s prevention and treatment, published in Nutrients.
I shared the findings from this literature review as a poster presenter at the Society for Neuroscience’s Global Connectome 2021.
Although clinical trials for VK2 in individuals with Alzheimer’s have not come to fruition, I was able to further investigate VK2 in a wet lab environment at North Central High School. My research on VK2’s effects on lifespan and gene expression in C. elegans earned me second and third-place awards in the Animal Sciences category at the Washington State Science & Engineering Fair.
From this project, I learned how to perform C. elegans culture and mass synchronization, lipid extraction from C. elegans, PCR, RT-qPCR, gel electrophoresis, library preparation for RNA-seq with Oxford Nanopore Technologies’ minION, and gene expression and pathway analysis.
Moreover, I was awarded the Biotechnician Assistant Credential (Credential ID 50488436) based on my performance on the BACE.
RSI Project conducted as part of the Rifkin Lab at UCSD
Gene regulatory networks, or GRNs, are critical for understanding evolutionary relationships and how organisms adapt to their environment. They provide a network-level view of the interactions occurring between DNA regulatory regions and transcription factors during gene regulation, and studies have shown that the organization of GRNs into groups, or modules, facilitates evolvability.
For my project, I constructed a simulation framework in C++ and within a virtual machine environment used evolutionary simulations involving different combinations of gene activity patterns to study how specialization, which involves selection for additional gene activity patterns, drives modularity.
My study demonstrated that specialization does indeed increase modularity, a finding generalizable to other types of networks. Developing a deeper network-level understanding of GRNs will enhance our ability to model the mechanisms underlying disease and discover biomarkers and therapeutics.
After learning about the serious bone loss problems associated with space travel that were preventing astronauts from going on longer space missions and the fact that our gut microbiome changes in space, I proposed a novel connection between bone health and the gut microbiome: Vitamin K2, which is produced by the bacteria in our gut.
I designed multiple experiments to test my hypothesis and presented my work to both subject-matter experts and the general public at the Finalist Launchpad.
Following the Launchpad, an abstract summarizing my research was published in the Journal of Emerging Investigators. The Genes in Space competition was the spark that ignited my fascination with the gut microbiome and Vitamin K2 and ultimately motivated my later research on Vitamin K2, including the literature review I coauthored the following year.

Featured in The Spokesman-Review: “NC sophomore has a date with NASA.” Here’s an out-of-this-world idea: A sophomore at North Central High School might help pave the way for astronauts to one day reach Mars.
During my first two years of high school, I participated in the BioBuilderClub, founded by MIT’s Natalie Kuldell to encourage high school student teams from around the world to design bioengineering projects using synthetic biology to address real-world problems.
Synthetic biology combines biology and engineering to model and construct useful living systems. Through synthetic biology, we try to genetically program cells to perform desirable tasks that ultimately advance our knowledge and help us find solutions to difficult problems.

My first project involved designing a biosensor for dichloromethane, a toxic pollutant present in a superfund site within a mile of my neighborhood that was leaking into groundwater and the Spokane Aquifer. My research was published in Biotreks.
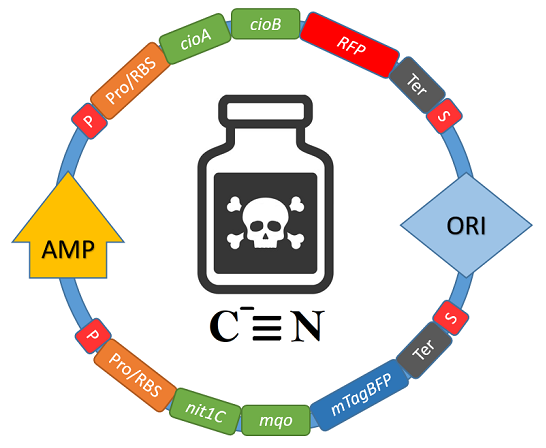
My second project was inspired by a story in my local newspaper about the presence of cyanide in our groundwater and involved designing a bioengineered bacterium to degrade this toxic substance. My research was again published in Biotreks and was recognized for scientific rigor and visual communications.
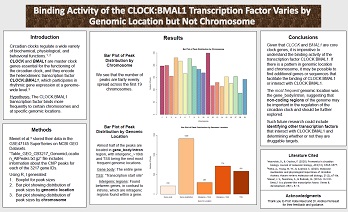
The importance of sleep in our health is not a new idea; however, there is emerging research on how circadian rhythms are disrupted in people with serious diseases.
When I attended and presented a poster on the K2/AD connection at the SfN Global Connectome, an international neuroscience conference, I learned that not only are there circadian disruptions in more than 80 percent of patients with Alzheimer’s disease, but also that the progression of Alzheimer’s has been associated with changes in the expression of clock genes.
I am interested in learning more about how gene expression varies with the environment and, specifically, about how circadian oscillations impact gene expression.
Under the guidance of Dr. Andrea Perreault and Katie Reed at Vanderbilt University, I studied how the binding activity of the CLOCK:BMAL1 transcription factor varies by genomic location and by chromosome. I wrote a scientific paper summarizing my work and presented my research at the Student Showcase of the conference “Genomics: The Next Generation of Biology Research.”










